Summary:
UCLA researchers in the Department of Radiological Sciences have developed a new computational pipeline for identifying lesions in MRI data.
Background:
Prostate cancer is one of the most common cancer types with 12.6% of men diagnosed within their lifetime. Prostate cancer ranks as the 2nd leading cause of cancer-related deaths in the United States and contributes significantly to Medicare expenditures, recently ranking as the highest mean expenditure. Increasingly, current radiology-based detection methods rely on multiple MRI sequences to grade the appearance of lesions based on the radiologist’s level of suspicion. However, the accuracy of this methodology is variable and highly dependent on the radiologist’s experience. This is largely due to benign elements which obfuscate cancerous lesions and result in a persistent need to confirm suspicious findings with biopsy, a costly and highly invasive procedure. Moreover, reliance on multiple MRI sequences leads to long patient scan times. Improved quantitative methods are needed to exploit subtle MRI imaging findings that may not be readily appreciated by the trained radiologist.
Innovation:
Dr. Yeager developed an algorithm that allows radiologists to identify statistically significant focal T2-intense regions whose global clustering patterns have been implicated as a defining characteristic of prostate cancer. With this algorithm and analysis pipeline, there is potential for improved cancer screening accuracy and reduced need for physical biopsy by improving the diagnostic sensitivity of MRI. This algorithm could simplify the current complex MRI prostate scanning protocols down to a single sequence and significantly reduce the total scan time per patient. Additionally, automation of this algorithm has the added potential to streamline the radiologist’s workload, diminishing the time spent to read each prostate study, as well as improving the radiologist’s consistency.
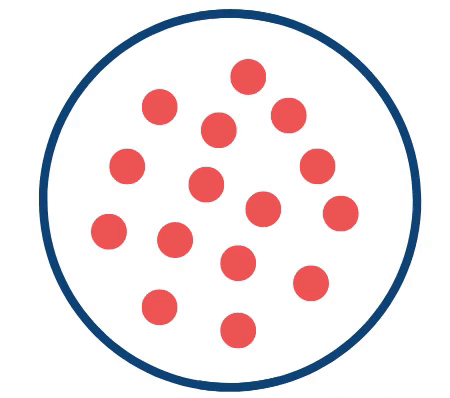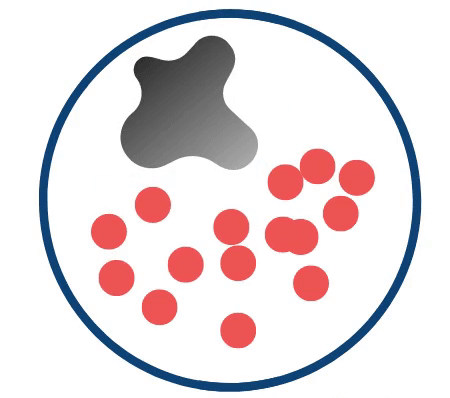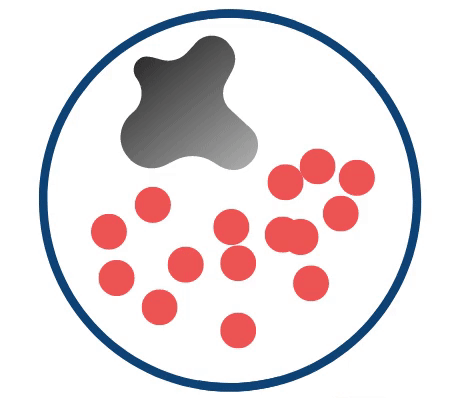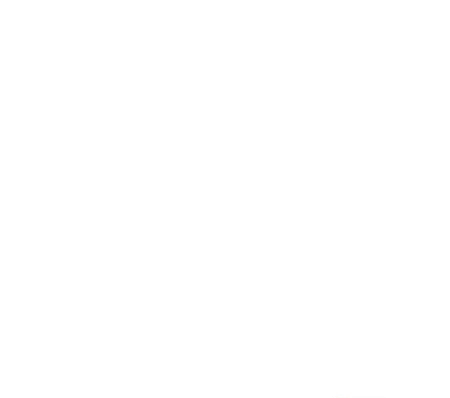
Credit: Yeager Lab. The above schematic highlights the prostate with T2-intense regions of interest (red circles) as they change in response to growing cancer (grey irregular shape).
Potential Applications:
• Prostate cancer identification
• Lesion characterization
Advantages:
• Simple and interpretable algorithm
• Characterizes pixel intensity distribution
• Extendable analysis to differentiate other non-cancerous lesions
• Reliance on a single MR sequence which directly translates to reduced MRI scan time
Development to Date:
A working pipeline has been developed for detecting benign and cancerous lesions based on a single MR sequence from data stored in the DICOM format.
Related Papers:
Gorman, B. L.; Yeager, A. N.; Chini, C. E.; Kraft, M. L. "Imaging the distributions of lipids and proteins in the plasma membrane with high-resolution secondary ion mass spectrometry," in Characterization of Biological Membranes: Structure and Dynamics; Eds.: Nieh, M.-P.; Heberle, F. A.; Katsaras, J.; De Gruyter: Berlin, Boston. 2019; Chapter 9, 287-322. DOI:10.1515/9783110544657-009.
Reference:
UCLA Case No. 2022-290
Lead Inventor:
Ashley Yeager