Summary:
UCLA researchers in the Departments of Bioengineering and Biological Chemistry have developed a method for identifying and enriching therapeutically useful cell sub-populations based on their secretions (SEC-seq).
Background:
Cell function is defined by a myriad of biomolecules that they secrete; over 3 thousand proteins are estimated to be secreted by human cells, spanning a diversity of critical functions. Currently, there are no available scalable methods to link individual cell secretions over time to their transcriptional state. Mesenchymal stromal cells (MSCs) have been widely evaluated for their use in cell therapies due to the favorable proteins they secrete. However, successful translations of cell therapies in MSCs have been hindered by clinical outcome inconsistency attributed to single-cell level functional differences within the cell population. Exploiting the full potential of cell therapies necessitates an understanding of the cell subpopulations that secrete specific proteins and how these secretomes change under the environments relevant to these therapies. Given the vast heterogeneity of relevant cell cultures, an ideal method would detect protein secretion at the single-cell level and link gene expression networks with those secretory behaviors. There remains a market need for a tool that can categorize gene expression at the single cell level for populations such as MSCs based on protein secretion.
Innovation:
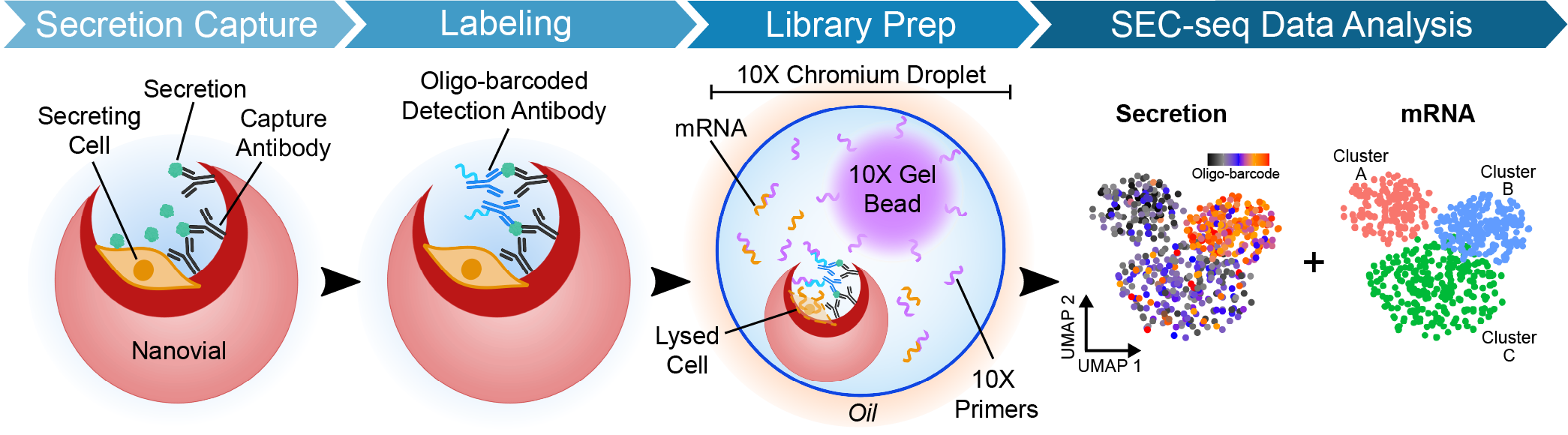
To address this gap, researchers in UCLA’s Di Carlo and Plath labs have developed secretion encoded single-cell sequencing (SEC-seq). This method utilizes functionalized hydrogel particles (nanovials) to capture individual cells, as well as their secretions, inside thousands of discrete reaction chambers. The SEC-seq workflow utilizes common laboratory analytical techniques, like flow cytometry and off-the-shelf sequencers, to screen for and analyze the desired subpopulations respectively. This method thus enables researchers to more easily and efficiently find MSC subpopulations with desirable secretomes for cell therapy applications. SEC-seq enables the identification of gene signatures linked to specific secretory states, enabling mechanistic studies, the isolation of secretory subpopulations, and the development of means to modulate cellular secretion. Using SEC-seq, one can identify surface markers that enable the purification and enrichment of highly-secreting MSC sub-populations. Ultimately, SEC-seq will enable a broader, democratized range of discoveries in cell biology and therapy development.
Video from the inventors: Nanovials: Technology created by UCLA Scientist Dino Di Carlo and colleagues aims to change research
Potential Applications:
• Identification of therapeutic cell populations (e.g., MSCs)
• Drug discovery and development
• Personalized medicine
• Mechanistic studies of protein secretion
• Multi-omics research
Advantages:
• Allows heterogeneity investigation
• Simultaneous measurement of secretion and transcriptome
• Compatible with common equipment, i.e. Flow cytometers and 10x Chromium
• High throughput and scalability
• Reduced crosstalk and enhanced specificity
State of Development:
Nanovials are manufactured and sold commercially, and SEC-seq has been demonstrated with MSCs and a corresponding manuscript has been published in a peer-reviewed journal.
Related Papers:
1. Udani, Shreya, et al. "Associating growth factor secretions and transcriptomes of single cells in nanovials using SEC-seq." Nature Nanotechnology 19.3 (2024): 354-363.
2. Koo, Doyeon, et al. "Defining T cell receptor repertoires using nanovial-based binding and functional screening." Proceedings of the National Academy of Sciences 121.14 (2024): e2320442121.
3. Langerman, J, et. al. "Linking single-cell transcriptomes with secretion using secretion-encoded single-cell sequencing (SEC-seq)" BioRxiv (2024)
Reference:
UCLA Case No. 2024-024
Lead Inventor:
Dino Di Carlo, UCLA Chair and Professor of Bioengineering.