Summary:
UCLA researchers have created a cutting-edge, automated phenotyping tool that harnesses electronic health records to enable rapid and precise identification of single ventricle physiology (SVP), especially in neonatal and pediatric populations.
Background:
Single ventricle physiology (SVP) is a rare heart condition that develops before birth and includes a spectrum of subtypes in which only one ventricle functions properly. SVP limits normal blood circulation due to the mixing of oxygenated and deoxygenated blood pumping through a single chamber of the heart. Due to its complexity and early onset in the body, medical professionals struggle to diagnose SVP, which may lead to delayed medical intervention and poor quality of life for newborns and infants. Current testing methods involve imaging and clinical assessment such as blood tests, ECG or EKG scans, echocardiograms, medical visits and research in case studies of individuals with SVP. However, these processes are time-consuming, resource-intensive, costly, inaccurate, and often inaccessible. Improving diagnostic accuracy and efficiency for SVP is critical to ensuring early intervention and significantly enhancing infant quality of life.
Innovation:
Professor Nguyen and her research team have developed an algorithm that identifies babies with SVP by analyzing clinical data in relation to international classification of disease (ICD) codes associated with the condition. This approach automates SVP diagnosis without the need for invasive procedures by drawing upon diverse data types from MRI scans to clinical notes. To develop the algorithm, a study of 1,020 infants was conducted and over 7 million electronic health records were obtained. The algorithm was then tested on 2,500 infants, with validation studies performed on an additional 22,500 infants. The result of these tests was a high accuracy yield of 91.18%. Benchmarking against published methods, the algorithm demonstrated 99.24% accuracy, 94.12% precision, 85.11% sensitivity, and an F1 score of 89.39%, a standard measure of classification model performance. Previous methods reported 95.20% accuracy, 43.23% precision, 88.30% sensitivity and 58.04% F1 score, reflecting a statistically significant improvement from the state of the art. Additionally, the algorithm facilitates the identification of false positives and negatives, improving overall diagnostic performance. The large sample size and high performance highlight the system’s potential to streamline medical procedures associated with SVP by precise and accurate diagnosis for improvement prognostication, monitoring and management.
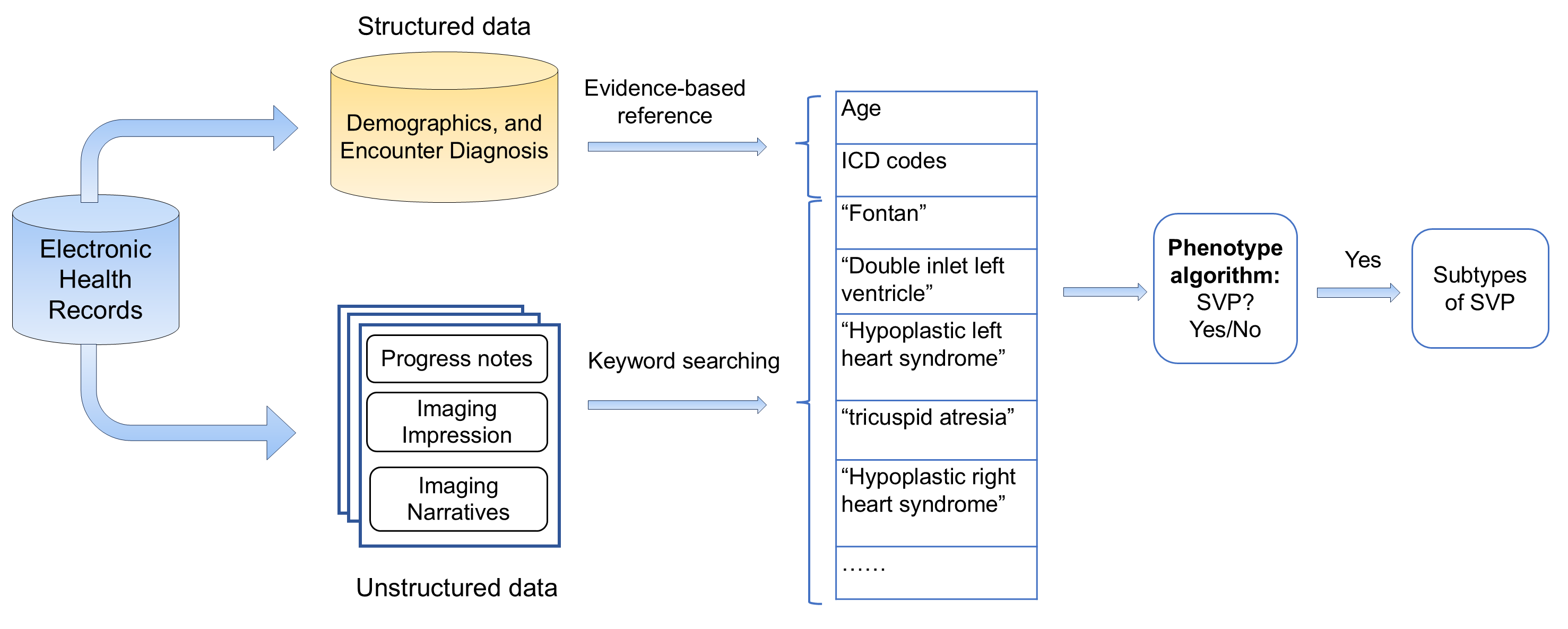
Source: Inventors' software Repository: https://github.com/Hang1205/A-phenotyping-algorithm-for-classification-of-single-ventricle-physiology-using-EHR
Potential Applications:
● Accurate SVP diagnosis
● Clinical case identification
● Automated clinical decision support
● Diagnostic support tools
● Resource planning for rare heart conditions
Advantages:
● Improves diagnostic precision
● Streamlines clinical workflows
● Enhances decision-making
● Reduces clinical misclassification
● Fully automated process
● Utilizes diverse data types
Status of Development:
The software code has been tested across a 2500 cohort to prove high accuracy, precision, and sensitivity.
Related Publications:
Xu H, Renella P, Badiyan R, Hindosh ZR, Elisarraras FX, Zhu B, Satou GM, Husain M, Finn JP, Hsu W, Nguyen KL. A phenotyping algorithm for classification of single ventricle physiology using electronic health records. JAMIA Open. 2025 May 15;8(3):ooaf035. doi: 10.1093/jamiaopen/ooaf035. PMID: 40376075; PMCID: PMC12080993.
Reference:
UCLA Case Nos. 2025-322 (software) and 2025-323
Lead Inventor:
Kim-Lien Nguyen, Faculty in the Department of Medicine